Müslüm Kaan Arıcı, Uncovering Hidden Connections and Functional Modules via pyPARAGON: A Hybrid Approach for Network Contextualization
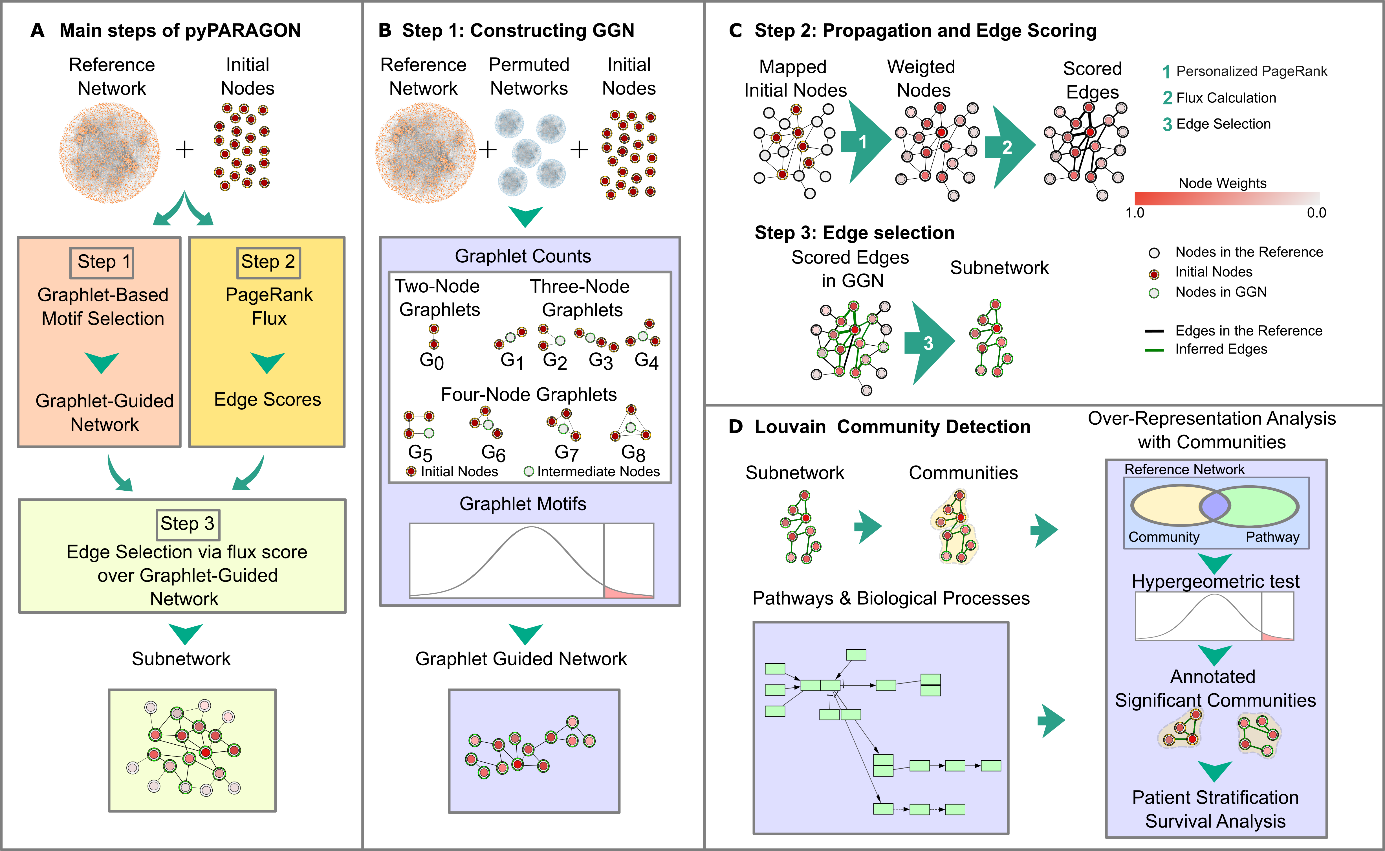
State-of-the-art omics technologies use network-based contextualization methods to give molecular information about different biological contexts, like disease states, patients, and drug changes. In the beginning, this thesis identified challenging issues such as missing points in contextualization, hidden knowledge in omics datasets, bias in reference networks, and noisy interactions with highly connected nodes or hubs. Subsequently, to address these challenges, we developed pyPARAGON (PAgeRAnk-flux on Graphlet-guided network for multi-Omics data integratioN). Also, a novel tool, pyPARAGON, contextualized patient datasets by inferring patient-specific networks and complex diseases by constructing disease models, namely breast cancer and autism spectrum disorders.
Date: 22.01.2024 / 14:00 Place: A-108